COCOA-Tree: COllaborative COevolution Analysis Toolbox including tree-based vizualisation¶
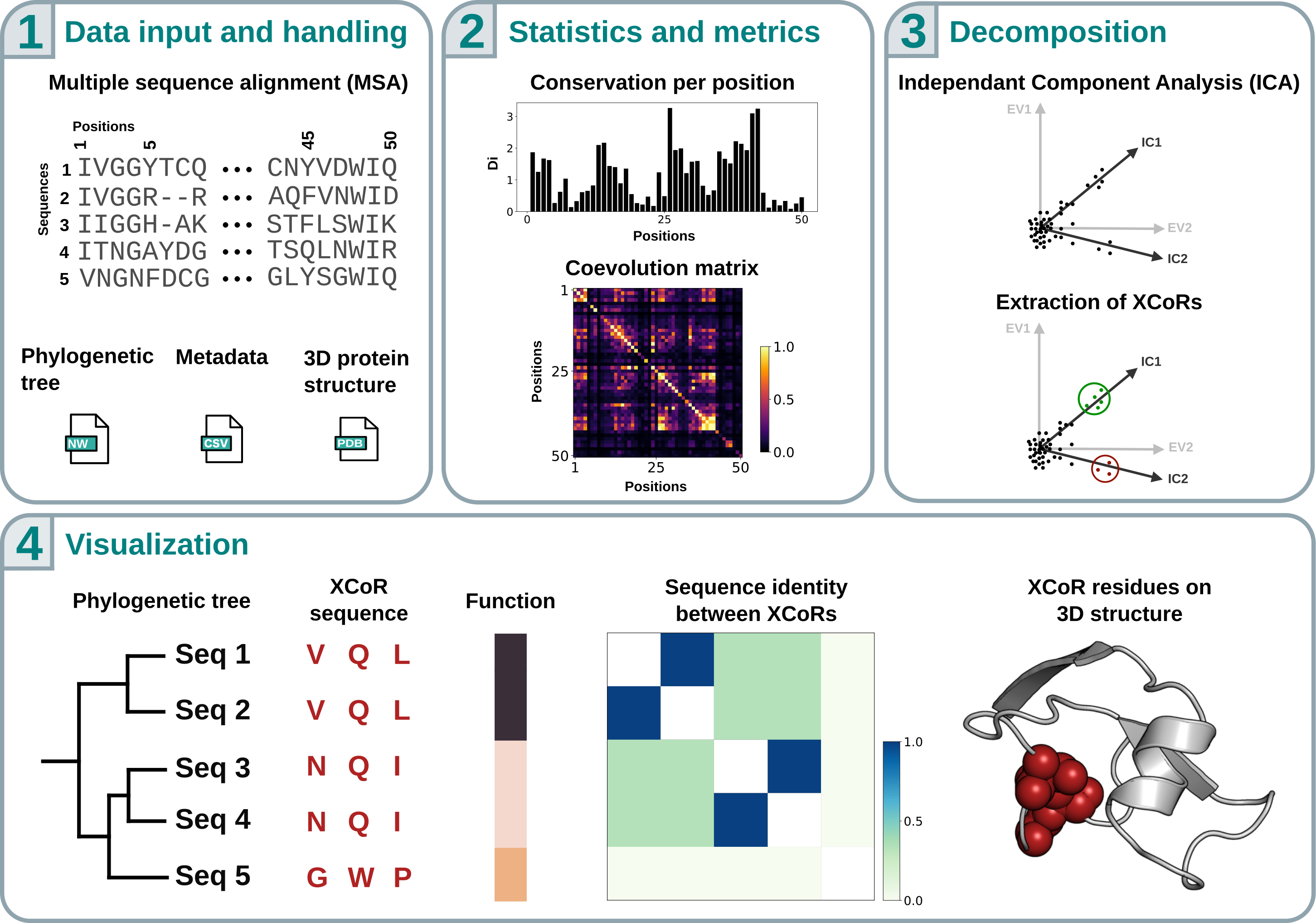
COCOA-Tree is a Python library designed to perform coevolution analysis of protein sequence data and to compare the results with phylogenetic trees and metadata. By doing so, it facilitates the investigation of the origins of coevolution patterns, their relationships to phylogeny and functional protein annotation, and ultimately aims to democratize the use and development of so-called protein sectors [RRR16] and specificity-determining positions (SDPs) [DJPV13].
The library includes several coevolution metrics (and allows users to define their own), along with various correction methods, enabling easy comparison between approaches. It also integrates with the molecular visualization software PyMOL, allowing users to map results onto 3D structures, more specifically, extremal co-evolving residues that form the basis of protein sectors [eal25].
The software is organized into different modules, detailed below:
List of abbreviations¶
Example gallery¶
Various examples of COCOA-Tree’s use can be found in the Gallery.
If you wish to perform a minimal SCA analysis, go to Minimal SCA example.
For a detailed SCA analysis, go to Full SCA analysis.
Citing COCOA-Tree¶
If you use COCOA-Tree in a scientific publication, we would appreciate citations to the following paper:
BEST PAPER EVER
Bibtex entry:
@article {}
License information¶
Conditions on the use and redistribution of this package.
Copyright (c) 2025 The COCOA-Tree developers.
All rights reserved.
Redistribution and use in source and binary forms, with or without
modification, are permitted provided that the following conditions are met:
1. Redistributions of source code must retain the above copyright notice,
this list of conditions and the following disclaimer.
2. Redistributions in binary form must reproduce the above copyright notice,
this list of conditions and the following disclaimer in the documentation
and/or other materials provided with the distribution.
3. Neither the name of the copyright holder nor the names of its contributors
may be used to endorse or promote products derived from this software without
specific prior written permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE
ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE
LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR
CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF
SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS
INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN
CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE)
ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF
THE POSSIBILITY OF SUCH DAMAGE.
David De Juan, Florencio Pazos, and Alfonso Valencia. Emerging methods in protein co-evolution. Nature Reviews Genetics, 14(4):249–261, April 2013. doi:10.1038/nrg3414.
Margaux et al. Tbd. BioRxiv, 1(1):1, 2025. doi:no DOI yet.
Najeeb Halabi, Olivier Rivoire, Stanislas Leibler, and Rama Ranganathan. Protein Sectors: Evolutionary Units of Three-Dimensional Structure. Cell, 138(4):774–786, August 2009. URL: https://linkinghub.elsevier.com/retrieve/pii/S0092867409009635 (visited on 2023-07-19), doi:10.1016/j.cell.2009.07.038.
Olivier Rivoire, Kimberly A. Reynolds, and Rama Ranganathan. Evolution-Based Functional Decomposition of Proteins. PLOS Computational Biology, 12(6):e1004817, June 2016. doi:10.1371/journal.pcbi.1004817.