Note
Go to the end to download the full example code. or to run this example in your browser via Binder
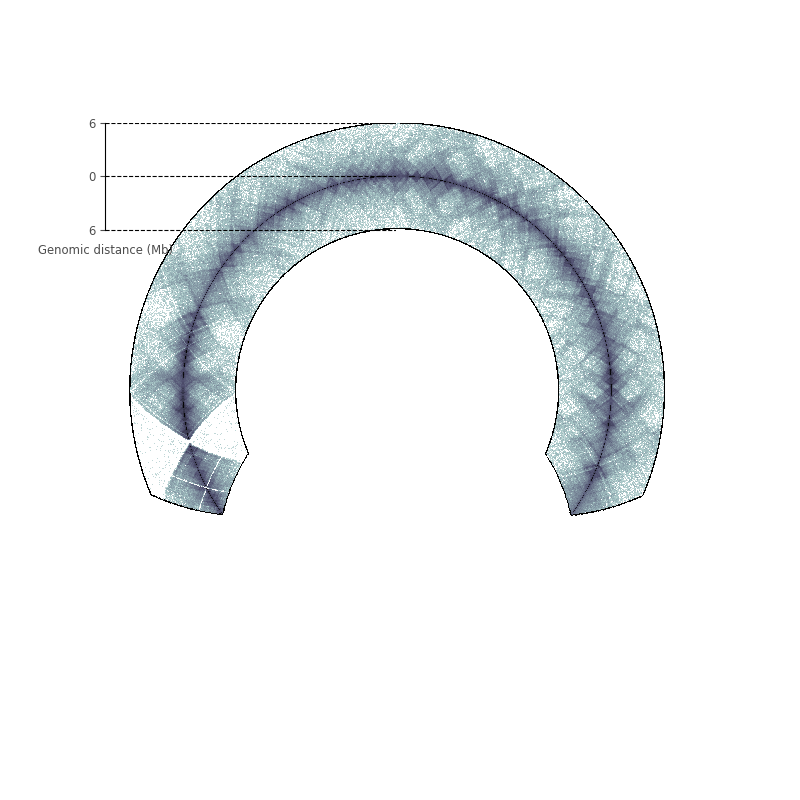
KBM-7 cell line, chromosome XI¶
Data: Rao SSP, et al. (2014) Cell 159(7):1665–1680
from circhic import datasets, CircHiCFigure
import matplotlib.pyplot as plt
data = datasets.load_kbm7()
counts = data["counts"]
nbins = data["nbins"]
circfig = CircHiCFigure(lengths=nbins, chromosome_type="linear")
circfig.plot_hic(counts, inner_gdis=120, outer_gdis=120, cmap="bone_r",
inner_radius=0.6, border_thickness=0.005)
rax = circfig.plot_raxis()
rax.set_yticklabels(["6", "0", "6"], fontsize="small")
rax.set_ylabel("Genomic distance (Mb)", fontsize="small", color="0.3",
position=(0, 1.03))
rax.tick_params(colors="0.3")
Total running time of the script: (0 minutes 3.697 seconds)