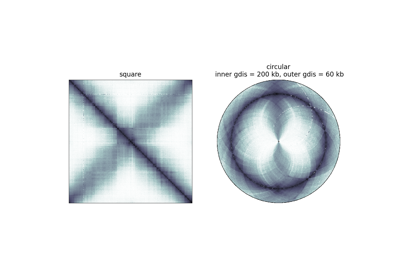
circhic.utils.generate_circular_map¶
- circhic.utils.generate_circular_map(data, granularity=0.5, inner_radius=0.5, resolution=None, inner_gdis=None, outer_gdis=None, mode='reflect', origin=1, chromosome_type='circular', frac_lin=0.7, rotate_lin=0)¶
Generate circular Strip Data
- Parameters:
- datandarray
Input data matrix to circularize with size (N, N)
- granularityfloat <= 1
granularityarity of display: the size of the output matrix is (Nc, Nc) where Nc=N/granularity; the smaller, the neater but also the longer
- inner_radiusfloat <= 1
Inner radius of the strip, supposing that the outer radius is equal to 1
- resolutioninteger, optional, default: None
resolution of data (in bp)
- inner_gdisinteger, optional, default: None
Genomic distance corresponding to the inner circle. By default, will be the maximum genomic distance when mode is “reflect”, and 0 if the mode is “distant”.
- outer_gdis: integer
Genomic distance corresponding to the outer circle. By default, will be the maximum genomic distance.
- mode{“reflect”, “distant”}, optional, default: “reflect”
if mode is “reflect”, the data will be ‘reflected’ around a genomic distance of 0, thus ranging from inner_gdis to 0 to outer_gdis. This equivalent to plotting both upper and lower triangular matrices of the original Hi-C contact count matrix.
if mode is “distant”, the data will be plotted from inner_gdis to outer_gdis.
- origin: integer
Genomic position at the vertical top. Only used if chromosome_type is “circular”.
- chromosome_typestring
‘circular’: circular chromosome, default ‘linear’: linear chromosome
- frac_linfloat <= 1
fraction of strip filled by linear chromosome
- rotate_linfloat <= 360
rotation angle (in degree) of the chromosome (the linear chromosome is symmetric with respect to the vertical axis by default)
- Returns:
- (Nc, Nc) ndarray containing the count data projected on a circular strip.