Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Square versus circular displays¶
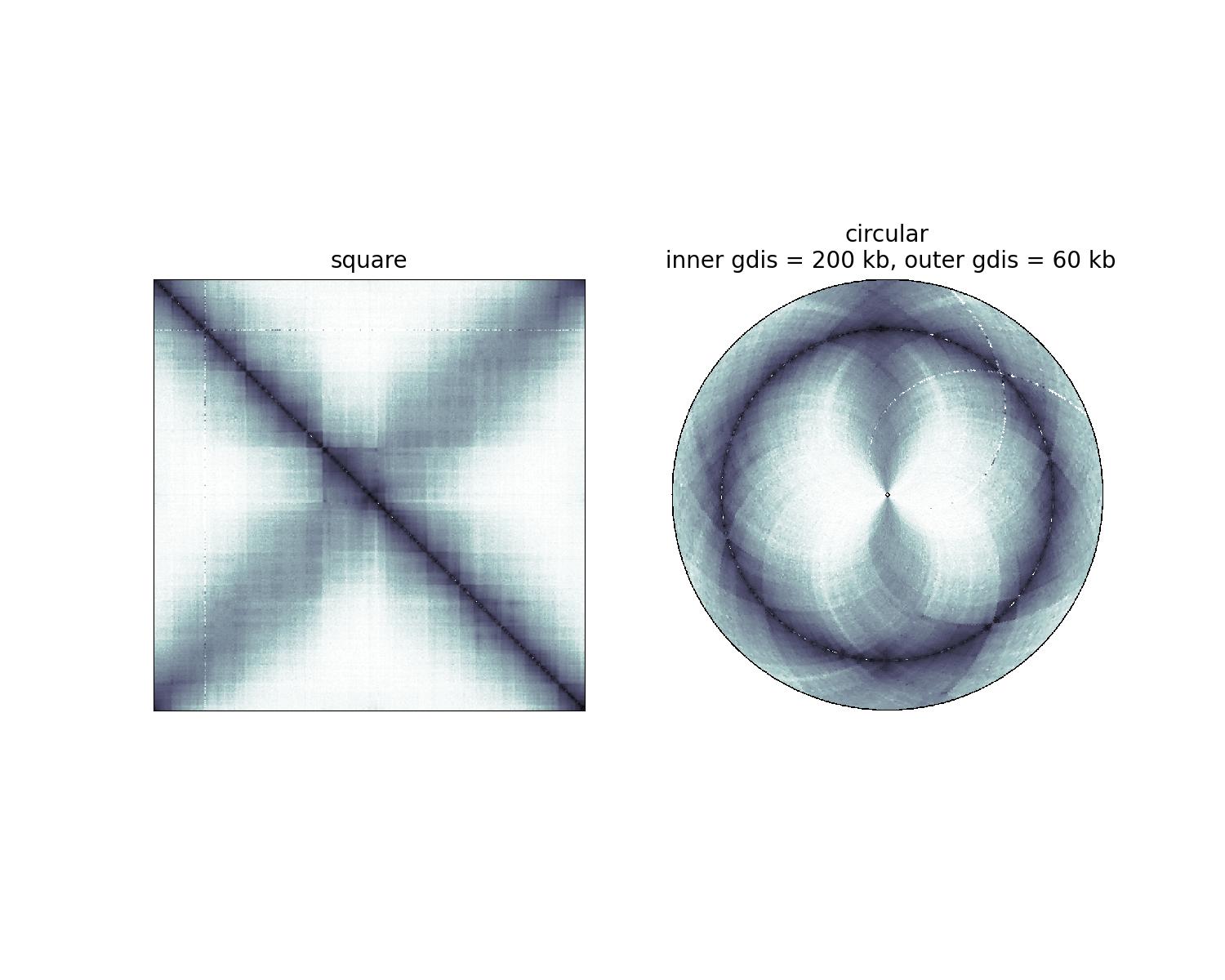
Text(0.5, 1.0, 'circular\n inner gdis = 200 kb, outer gdis = 60 kb')
import numpy as np
import matplotlib # Need this to check the version
import matplotlib.pyplot as plt
from matplotlib import colors
from circhic import datasets
from circhic import CircHiCFigure
from circhic.utils import generate_circular_map
from circhic.utils import generate_borders
from iced.normalization import ICE_normalization
#load the data
data = datasets.load_ccrescentus()
counts = data["counts"]
lengths = data["nbins"]
counts, bias = ICE_normalization(counts, output_bias=True)
granularity = 0.5
inner_radius = 0.005
resolution = 9958
inner_gdis, outer_gdis = 2000000, 600000
Circ = generate_circular_map(counts, granularity=granularity,
inner_radius=inner_radius, resolution=resolution,
inner_gdis=inner_gdis, outer_gdis=outer_gdis,
mode='reflect', origin=1,
chromosome_type='circular')
Bord = generate_borders(counts, granularity=granularity,
inner_radius=inner_radius, resolution=resolution,
inner_gdis=inner_gdis, outer_gdis=outer_gdis, origin=1,
chromosome_type='circular', thick_r=0.006)
vmax = np.max([counts[i, (i+1) % lengths[0]] for i in range(lengths[0])])
vmin = 77
plt.figure(figsize=(15, 12))
plt.subplot(1, 2, 1)
if matplotlib.__version__ < "3.2.0":
norm = colors.SymLogNorm(vmin, vmax=vmax)
else:
norm = colors.SymLogNorm(vmin, vmax=vmax, base=np.e)
plt.imshow(counts, norm=norm, cmap='bone_r')
plt.xticks([])
plt.yticks([])
plt.title('square', pad=10, fontsize=20)
plt.subplot(1, 2, 2)
plt.imshow(Circ, norm=norm, cmap='bone_r',
interpolation='None')
plt.imshow(vmax*Bord, norm=norm, cmap='Greys',
interpolation='None')
plt.xticks([])
plt.yticks([])
plt.axis('off')
plt.title('circular\n inner gdis = 200 kb, outer gdis = 60 kb', pad=10,
fontsize=20)
Total running time of the script: (0 minutes 1.016 seconds)