Note
Go to the end to download the full example code. or to run this example in your browser via Binder
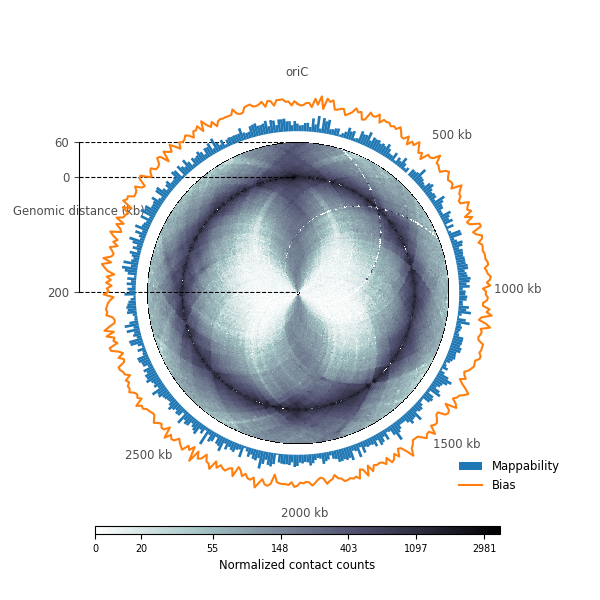
Circular Hi-C and genomic data¶
/usr/share/miniconda3/envs/circhic/lib/python3.8/site-packages/iced/normalization/_ca_utils.py:8: UserWarning: The API of this module is likely to change. Use only for testing purposes
warnings.warn(
import numpy as np
import matplotlib.pyplot as plt
from circhic import datasets
from circhic import CircHiCFigure
from iced.normalization import ICE_normalization
# Load the data, compute the cumulative raw counts.
data = datasets.load_ccrescentus()
counts_raw = data["counts"]
lengths = data["nbins"]
cumul_raw_counts = counts_raw.sum(axis=0)
# Normalize the data using ICE, and keep the biases
counts, bias = ICE_normalization(counts_raw, output_bias=True)
# compute extreme values
vmax = np.max([counts[i, (i+1) % counts.shape[0]]
for i in range(counts.shape[0])])
vmin = np.min(counts[counts > 0])
# plotting the data
granularity = 0.5
resolution = 9958
fig = plt.figure(figsize=(6, 6))
inner_radius, outer_radius = 0.005, 0.75
inner_gdis, outer_gdis = 2000000, 600000
chrom_lengths = lengths * resolution
circhicfig = CircHiCFigure(chrom_lengths, figure=fig)
m, ax = circhicfig.plot_hic(counts, granularity=granularity,
resolution=resolution, outer_radius=outer_radius,
inner_radius=inner_radius, inner_gdis=inner_gdis,
outer_gdis=outer_gdis, vmin=vmin*100, vmax=vmax,
cmap="bone_r", border_thickness=0.005)
rax = circhicfig.plot_raxis()
rax.set_yticklabels(["200", "0", "60"], fontsize="small")
rax.set_ylabel("Genomic distance (kb)", fontsize="small", color="0.3",
position=(0, 1.03))
rax.tick_params(colors="0.3")
# Assume you want to plot data from that ranges in a polar plot outside of the
# first one. Then the 0 axis should be at, say, 80% of the axis
bar, _ = circhicfig.plot_bars(
cumul_raw_counts, inner_radius=0.8, outer_radius=0.9,
color="C0")
# Now, plot another plot, for the top 10% of the original axes
lines, _ = circhicfig.plot_lines(
bias, color="C1", inner_radius=0.9, outer_radius=1)
cab = circhicfig.set_colorbar(m, orientation="horizontal")
cab.set_label("Normalized contact counts", fontsize="small")
ticklabels = ["%d kb" % (i * 500) for i in range(6)]
tickpositions = [int(i*500000) for i in range(6)]
ticklabels[0] = "oriC"
ax = circhicfig.set_genomic_ticklabels(
tickpositions=tickpositions,
ticklabels=ticklabels,
outer_radius=1, fontdict={'fontsize': "small"})
ax.tick_params(colors="0.3")
fig.legend((bar, lines[0]), ("Mappability", "Bias"), fontsize="small",
bbox_to_anchor=(0.8, 0.1, 0.15, 0.15), frameon=False)
print("")
Total running time of the script: (0 minutes 1.493 seconds)