cocoatree.deconvolution.extract_independent_components¶
- cocoatree.deconvolution.extract_independent_components(coevo_matrix, method=None, n_components=3, nrandom_pySCA=10, sequences=None, learnrate_ICA=0.1, nb_iterations_ICA=100000, freq_regul=0.03, verbose_random_iter=True)[source]¶
Extract independent components from a coevolution matrix
The current method is fully applicable to SCA analysis. For other metrics, we set n_components = 3 (to improve)
Parameters¶
- coevo_matrixnp.ndarray
coevolution matrix
- sequenceslist of sequences, optional, default: None
when using pySCA’s strategy to estimate the number of components, sequences needs to be provided.
- method{None, “pysca”}, default=None
Methods to use to estimate the number of components to extract. By default, relies on the number of components provided by the user.
- n_componentsint, default=3,
Number of independent components to extract
- nrandom_pySCAint, default=10,
Number of MSA randomizations to perform if method=’pySCA’
- learnrate_ICAint, default=0.1,
Learning rate / relaxation parameter used if method=’pySCA’
- nb_iteration_ICAint, default=100000,
Number of iterations if method=’pySCA’
freq_regul : regularization parameter (default=__freq_regularization_ref)
verbose_random_iter : Boolean
Returns¶
- idpt_componentsndarray of shape (n_components, n_pos)
corresponding to a list of independent components
Examples using cocoatree.deconvolution.extract_independent_components¶
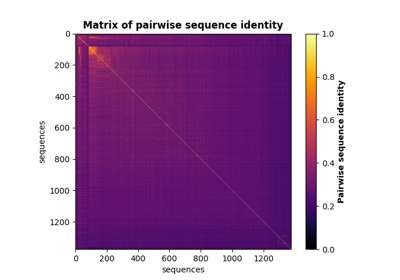
Perform full SCA analysis on the S1A serine protease dataset