cocoatree.msa.compute_seq_identity¶
- cocoatree.msa.compute_seq_identity(sequences)[source]¶
Computes the identity between sequences in a MSA (as Hamming’s pairwise distance)
Parameters¶
sequences : list of sequences
Returns¶
sim_matrix : identity matrix of shape (Nseq, Nseq)
Examples using cocoatree.msa.compute_seq_identity¶
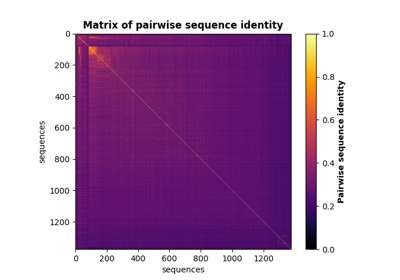
Perform full SCA analysis on the S1A serine protease dataset
Perform full SCA analysis on the S1A serine protease dataset
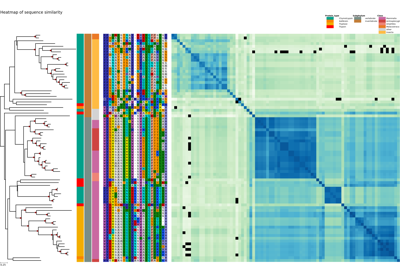
Plot a similarity heatmap of a XCoR along the phylogenetic tree
Plot a similarity heatmap of a XCoR along the phylogenetic tree