cocoatree.perform_sca¶
- cocoatree.perform_sca(sequences_id, sequences, n_components=4, freq_regul=0.03, gap_threshold=0.4, seq_threshold=0.2, coevolution_metric='SCA', correction=None)[source]¶
Perform statistical coupling analysis (SCA)
Parameters¶
sequences : list of MSA sequences to filter
sequences_id : list of the MSA’s sequence identifiers
n_components : int, default: 4
- gap_thresholdfloat [0, 1], default: 0.4
max proportion of gaps tolerated
seq_threshold : maximum fraction of gaps per sequence (default 0.2)
- coevolution_metricstr or callable, optional, default: ‘SCA’
which coevolution metric to use:
SCA: the coevolution matrix from Rivoire et al
MI: the mutual information
NMI: the normalized mutual information
callable: a function that takes as arguments (1) sequences, (2) seq_weights, and freq_regul
- correction{None, ‘APC’, ‘entropy’}, default: None
which correction to use
Returns¶
- coevol_matrixnp.ndarray (n_filtered_pos, n_filtered_pos)
coevolution matrix
- coevol_matrix_ngmnp.ndarray (n_filtered_pos, n_filtered_pos)
coevolution matrix without global mode (ngm = no global mode)
df : pd.DataFrame with the following columns
original_msa_pos : the original MSA position
filtered_msa_pos : the position in the filtered MSA
and for each component:
PCk: the projection of the residue onto the kth principal component
ICk: the projeciton of the residue onto the kth independent component
xcor_k: wherether the residue is found to be part of xcor k
Examples using cocoatree.perform_sca¶
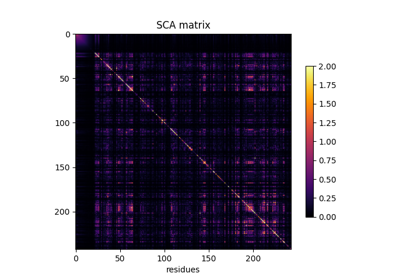
A minimal coevo analysis using cocoatree.perform_sca()