Note
Go to the end to download the full example code. or to run this example in your browser via Binder
The simplest SCA analysis ever¶
This example shows the full process to perform a complete SCA analysis and detect protein sectors from data importation, MSA filtering.
import cocoatree.datasets as c_data
import cocoatree
import matplotlib.pyplot as plt
Load the dataset¶
We start by importing the dataset. In this case, we can directly load the S1
serine protease dataset provided in cocoatree. To work on your on
dataset, you can use the cocoatree.io.load_msa function.
serine_dataset = c_data.load_S1A_serine_proteases()
loaded_seqs = serine_dataset["alignment"]
loaded_seqs_id = serine_dataset["sequence_ids"]
n_loaded_pos, n_loaded_seqs = len(loaded_seqs[0]), len(loaded_seqs)
Compute the SCA analysis¶
coevol_matrix, results = cocoatree.perform_sca(
loaded_seqs_id, loaded_seqs, n_components=3)
print(results.head())
computing weight of seq 1/1376
computing weight of seq 101/1376
computing weight of seq 201/1376
computing weight of seq 301/1376
computing weight of seq 401/1376
computing weight of seq 501/1376
computing weight of seq 601/1376
computing weight of seq 701/1376
computing weight of seq 801/1376
computing weight of seq 901/1376
computing weight of seq 1001/1376
computing weight of seq 1101/1376
computing weight of seq 1201/1376
computing weight of seq 1301/1376
original_msa_pos filtered_msa_pos PC1 IC1 ... sector_2 PC3 IC3 sector_3
0 0 NaN NaN NaN ... False NaN NaN False
1 1 NaN NaN NaN ... False NaN NaN False
2 2 NaN NaN NaN ... False NaN NaN False
3 3 NaN NaN NaN ... False NaN NaN False
4 4 NaN NaN NaN ... False NaN NaN False
[5 rows x 11 columns]
Select the position of the first sector in the results dataframe.
print(results.loc[results["sector_1"]].head())
original_msa_pos filtered_msa_pos PC1 ... PC3 IC3 sector_3
130 130 23.0 -0.090217 ... 0.029506 0.045516 False
133 133 26.0 -0.116480 ... 0.044673 0.059013 False
214 214 35.0 -0.106719 ... -0.087057 -0.046816 False
246 246 48.0 -0.130522 ... -0.028971 -0.038197 False
247 247 49.0 -0.110328 ... 0.038413 0.023443 False
[5 rows x 11 columns]
Visualizing the sectors on the first and second PC¶
# Plotting all elements in components
fig, ax = plt.subplots()
ax.plot(results.loc[:, "PC1"],
results.loc[:, "PC2"],
".", c="black")
# Plotting elements in sectors
for isec, color in zip([1, 2, 3], ['r', 'g', 'b']):
ax.plot(results.loc[results["sector_%d" % isec], "PC1"],
results.loc[results["sector_%d" % isec], "PC2"],
".", c=color, label="Sector %d" % isec)
ax.set_xlabel("PC1")
ax.set_ylabel("PC2")
ax.legend()
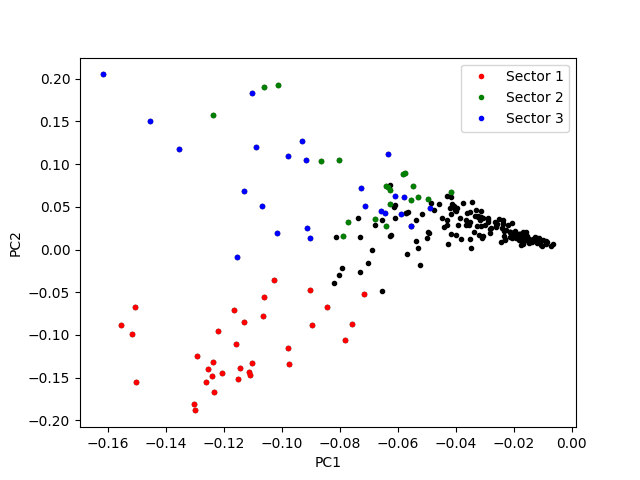
<matplotlib.legend.Legend object at 0x7fc006207c10>
Visualizing the sectors on the first and second IC¶
# Plotting all elements in components
fig, ax = plt.subplots()
ax.plot(results.loc[:, "IC1"],
results.loc[:, "IC2"],
".", c="black")
# Plotting elements in sectors
for isec, color in zip([1, 2, 3], ['r', 'g', 'b']):
ax.plot(results.loc[results["sector_%d" % isec], "IC1"],
results.loc[results["sector_%d" % isec], "IC2"],
".", c=color, label="Sector %d" % isec)
ax.set_xlabel("IC1")
ax.set_ylabel("IC2")
ax.legend()
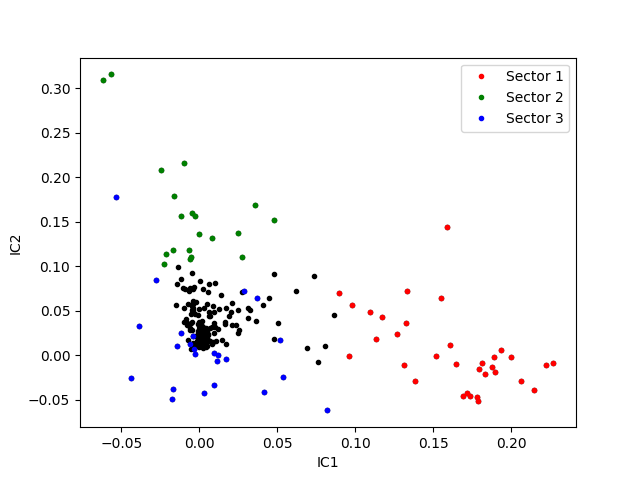
<matplotlib.legend.Legend object at 0x7fc005eecd10>
Total running time of the script: (0 minutes 2.549 seconds)