Note
Go to the end to download the full example code or to run this example in your browser via Binder.
A minimal coevo analysis¶
This example shows how to perform a minimal co-evolutionary analysis using the SCA approach. This includes:
generating a SCA co-evolutionary matrix, reduce it, and sort it according
to XCoRs,
showing the position composition of XCoRs,
plotting the XCoRs using the independent component projections.
# Author: Margaux Jullien <margaux.jullien@univ-grenoble-alpes.fr>
# Nelle Varoquaux <nelle.varoquaux@univ-grenoble-alpes.fr>
# Ivan Junier <ivan.junier@univ-grenoble-alpes.fr>
# License: TBD
Necessary imports
import numpy as np
import matplotlib.pyplot as plt
import cocoatree.datasets as c_data
import cocoatree.msa as c_msa
import cocoatree.decomposition as c_decomp
from cocoatree.statistics.pairwise import compute_sca_matrix
Loading the dataset¶
serine_dataset = c_data.load_S1A_serine_proteases()
loaded_seqs = serine_dataset["alignment"]
loaded_seqs_id = serine_dataset["sequence_ids"]
n_loaded_pos, n_loaded_seqs = len(loaded_seqs[0]), len(loaded_seqs)
Filtering the MSA¶
Computing the SCA matrix¶
Plotting the SCA matrix¶
fig, ax = plt.subplots()
im = ax.imshow(SCA_matrix, vmin=0, vmax=2, cmap='inferno')
ax.set_xlabel('residues', fontsize=10)
ax.set_ylabel(None)
ax.set_title('SCA matrix')
fig.colorbar(im, shrink=0.7)
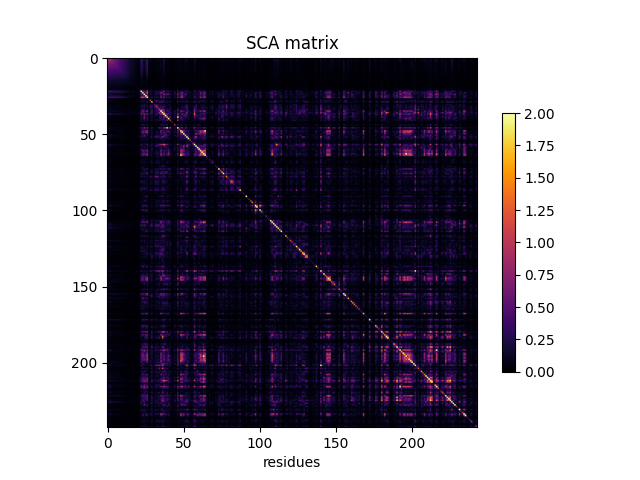
<matplotlib.colorbar.Colorbar object at 0x7f61e4fdea10>
Extracting XCoRs¶
XCoR positions on (filtered) sequences:
XCoR_1: [64, 48, 63, 195, 182, 168, 197, 199, 108, 61, 144, 146, 198, 196, 216, 49, 192, 194, 222, 234, 145, 208, 211, 35, 235, 26, 62, 50, 155, 200, 97, 23, 73]
XCoR_2: [140, 202, 52, 57, 110, 111, 129, 78, 87, 58, 226, 75, 124, 128, 81, 34, 231, 32, 33, 204]
XCoR_3: [212, 225, 190, 180, 223, 183, 213, 210, 193, 224, 220, 172, 184, 176, 36, 217, 161, 37, 142, 160, 177, 46, 188, 24]
A plotting function¶
def plot_coevo_according2xcors(coevo_matrix, xcors=[], vmin=0, vmax=1e6):
""" A plotting function returning a reduced coevo matrix, keeping
only XCoRs and sorting the residues according to them
"""
fig, ax = plt.subplots(tight_layout=True)
xcor_sizes = [len(x) for x in xcors]
cumul_sizes = sum(xcor_sizes)
sorted_pos = [p for xcor in xcors for p in xcor]
im = ax.imshow(coevo_matrix[np.ix_(sorted_pos, sorted_pos)],
vmin=vmin, vmax=vmax,
interpolation='none', aspect='equal',
extent=[0, cumul_sizes, cumul_sizes, 0],
cmap='inferno')
cb = fig.colorbar(im)
cb.set_label("coevolution level")
line_index = 0
for i in range(n_xcors):
ax.plot([line_index + xcor_sizes[i], line_index + xcor_sizes[i]],
[0, cumul_sizes], 'w', linewidth=2)
ax.plot([0, cumul_sizes],
[line_index + xcor_sizes[i], line_index + xcor_sizes[i]],
'w', linewidth=2)
line_index += xcor_sizes[i]
ticks = []
for ix in range(len(xcors)):
shift = np.sum([len(xcors[j]) for j in range(ix)])
ticks.append(shift+len(xcors[ix])/2)
ax.set_xticks(ticks)
ax.set_xticklabels(['XCoR_%d' % ix for ix in range(1, len(xcors)+1)])
ax.set_yticks(ticks)
ax.set_yticklabels(['XCoR_%d' % ix for ix in range(1, len(xcors)+1)],
rotation=90, va='center')
return fig, ax
Plotting the SCA matrix according to the XCoRs¶
fig, ax = plot_coevo_according2xcors(SCA_matrix, xcors, vmin=0, vmax=2)
ax.set_title('SCA matrix, sorted according to XCoRs')
ax.set_xlabel('XCoR\'s positions', fontsize=10)
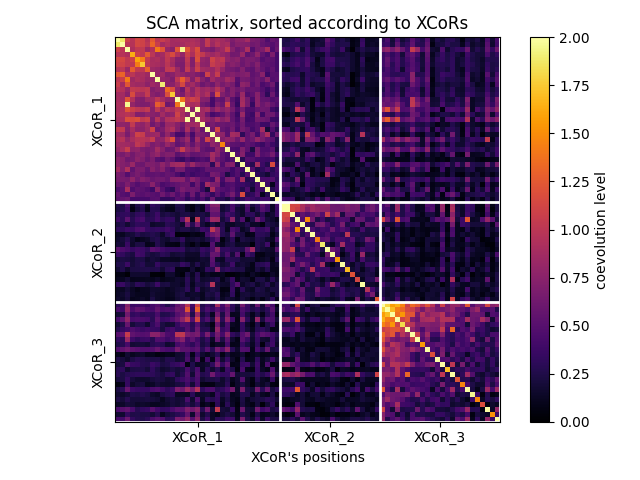
Text(0.5, 29.000000000000014, "XCoR's positions")
Removing a global mode (ngm = no global mode), i.e., setting largest eigeinvalue to zero
Plotting the SCA matrix without global mode according to the XCoRs¶
fig, ax = plot_coevo_according2xcors(SCA_matrix_ngm, xcors, vmin=0, vmax=2)
ax.set_title('SCA matrix without global mode\nsorted according to XCoRs')
ax.set_xlabel('XCoR\'s positions', fontsize=10)

Text(0.5, 29.000000000000014, "XCoR's positions")
Adapting the coevolution scale¶
fig, ax = plot_coevo_according2xcors(SCA_matrix_ngm, xcors, vmin=0, vmax=1)
ax.set_title('SCA matrix without global mode\nsorted according to XCoRs')
ax.set_xlabel('XCoR\'s positions', fontsize=10)
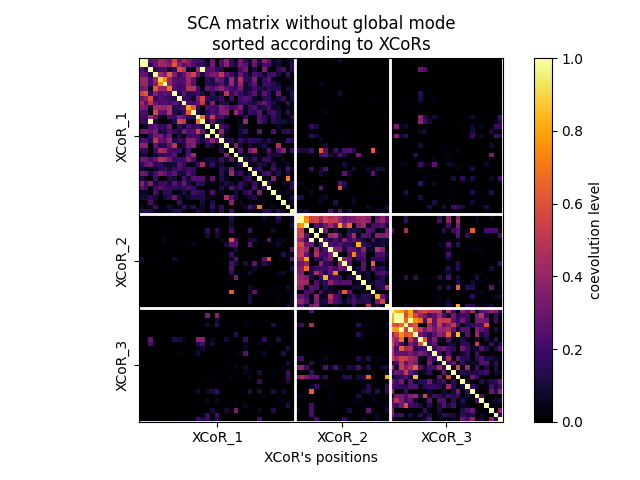
Text(0.5, 29.000000000000014, "XCoR's positions")
Visualizing XCoRs on independent components¶
# extracting independent components
ICs = c_decomp.extract_independent_components(SCA_matrix,
n_components=n_xcors)
# plotting IC_1 values versus IC_2 values
fig, ax = plt.subplots()
ax.plot(ICs[0], ICs[1], '.k')
# highlighting XCoR-associated values using a color code
# red: XCoR_1, green: XCoR_2, blue: XCoR_3
for ixcor, color in zip([0, 1, 2], ['r', 'g', 'b']):
ax.plot(ICs[0][xcors[ixcor]], ICs[1][xcors[ixcor]],
'.', c=color, label='XCoR_%d' % (ixcor+1))
ax.set_xlabel('IC1')
ax.set_ylabel('IC2')
ax.legend()
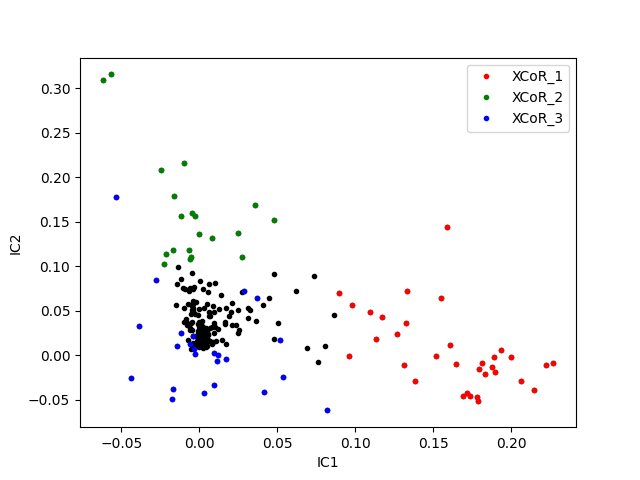
<matplotlib.legend.Legend object at 0x7f61e4e72290>
Total running time of the script: (0 minutes 2.761 seconds)