cocoatree.msa.filter_sequences¶
- cocoatree.msa.filter_sequences(sequences, sequences_id, gap_threshold=0.4, seq_threshold=0.2, verbose=False)[source]¶
Filter sequences
Remove (1) overly gapped positions; (2) overly gapped sequences.
Parameters¶
sequences : list of MSA sequences to filter
sequences_id : list of the MSA’s sequence identifiers
- gap_thresholdfloat,
maximum proportion of gaps tolerated per position (default=0.4)
- seq_thresholdfloat,
maximum proportion of gaps tolerated per sequence (default=0.2)
Returns¶
- filtered_seqslist of the remaining sequences (written as strings)
after applying the filters
- filtered_seqs_idlist of sequence identifiers that were kept after
applying the filters
- remaining_posnumpy.ndarray
remaining positions after filtering
Examples using cocoatree.msa.filter_sequences¶
Mapping original MSA, filtered MSA, PDB, and XCoRs
Mapping original MSA, filtered MSA, PDB, and XCoRs
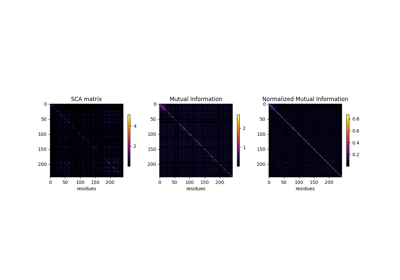
Mutual information versus SCA co-evolution metrics
Mutual information versus SCA co-evolution metrics
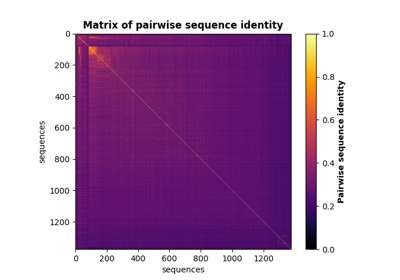
Perform full SCA analysis on the S1A serine protease dataset
Perform full SCA analysis on the S1A serine protease dataset