cocoatree.datasets.load_S1A_serine_proteases¶
- cocoatree.datasets.load_S1A_serine_proteases(paper='rivoire')[source]¶
Load the S1A serine protease dataset
Halabi dataset: 1470 sequences of length 832; 3 sectors identified Rivoire dataset : 1390 sequences of length 832 (snake sequences were removed for the paper’s analysis); 6 sectors identified (including the 3 from Halabi et al, 2008)
Parameters¶
- paper: str, either ‘halabi’ or ‘rivoire’
whether to load the dataset from Halabi et al, Cell, 2008 or from Rivoire et al, PLoS Comput Biol, 2016
Returns¶
- a dictionnary containing :
sequences_ids: a list of strings corresponding to sequence names
alignment: a list of strings corresponding to sequences. Because it is an MSA, all the strings are of same length.
metadata: a pandas dataframe containing the metadata associated with the alignment.
sector_positions: a dictionnary of arrays containing the residue positions associated to each sector, either in Halabi et al, or in Rivoire et al.
pdb_sequence: sequence extracted from rat’s trypsin PDB structure
pdb_positions: positions extracted from rat’s trypsin PDB structure
Examples using cocoatree.datasets.load_S1A_serine_proteases¶
Mapping original MSA, filtered MSA, PDB, and XCoRs
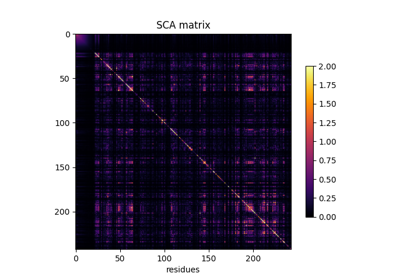
A minimal coevo analysis using cocoatree.perform_sca()
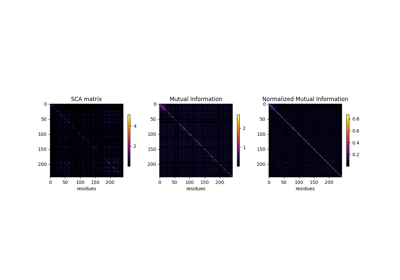
Mutual information versus SCA co-evolution metrics
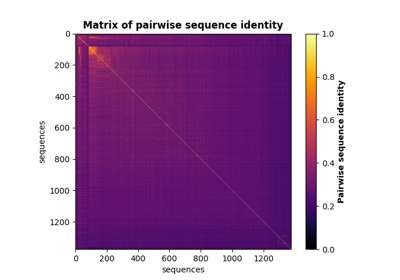
Perform full SCA analysis on the S1A serine protease dataset
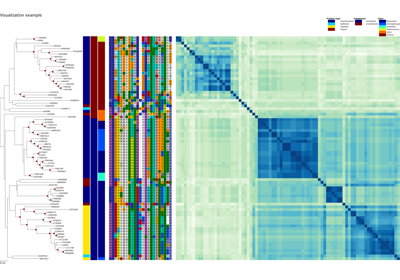
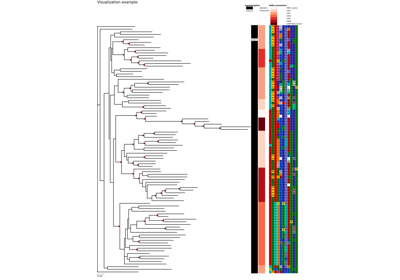
Plot XCoR together with (phylogenetic) tree and metadata
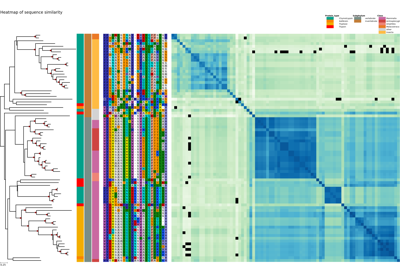
Plot a similarity heatmap of a XCoR along the phylogenetic tree