cocoatree.statistics.position.compute_conservation¶
- cocoatree.statistics.position.compute_conservation(sequences, seq_weights=None, freq_regul=0.03)[source]¶
Compute the conservation of amino acid at each position.
The conservation is computed as the relative entropy (e.g., the Kullback-Leibler divergence)
\[D_i^a = f_i^a \ln \frac{f_i^a}{q^a} + (1 - f_i^a) \ln \frac{1 - f_i^a}{1 - q^a}\]- where \(f_i^a\) is the observed frequency of amino acid a at
position i`, \(q^a\) is the background expectation
\(D_i^a\) indicates how unlikely the observed frequencies of amino acid a at position i would be if a occurred randomly with probability \(q^a\).
Parameters¶
sequences : list of sequences
- seq_weightsndarray (nseq), optional, default: None
if None, will compute sequence weights
freq_regul : regularization parameter (default=__freq_regularization_ref)
Returns¶
- Dinp.ndarray (npos,)
where each entry corresponds to the conservation at this position in the sequences.
Examples using cocoatree.statistics.position.compute_conservation¶
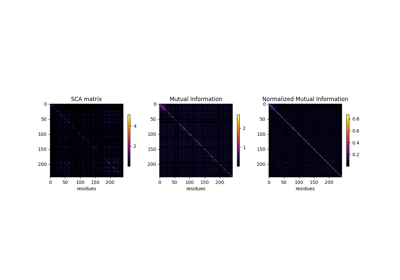
Mutual information versus SCA co-evolution metrics
Mutual information versus SCA co-evolution metrics
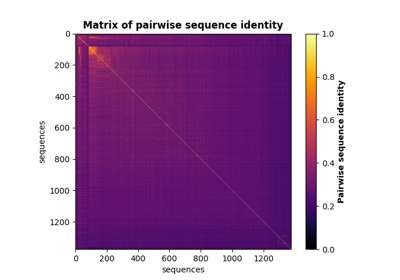
Perform full SCA analysis on the S1A serine protease dataset
Perform full SCA analysis on the S1A serine protease dataset