Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Mutual information versus SCA co-evolution metrics¶
In this example, we are comparing the results of the co-evolution analysis on serine proteases using SCA and the mutual information.
Necessary imports
from cocoatree.datasets import load_S1A_serine_proteases
from cocoatree.msa import filter_sequences
from cocoatree.statistics.position import compute_conservation
from cocoatree.statistics.pairwise import compute_sca_matrix
from cocoatree.statistics.pairwise import compute_mutual_information_matrix
import matplotlib.pyplot as plt
import numpy as np
Load the dataset¶
We start by importing the dataset.
For more details on the S1A serine proteases dataset, go to S1A serine proteases.
serine_dataset = load_S1A_serine_proteases()
seq_id = serine_dataset["sequence_ids"]
sequences = serine_dataset["alignment"]
n_pos, n_seq = len(sequences[0]), len(sequences)
Filtering of the multiple sequence alignment¶
We filter and clean the MSA
Compute the SCA matrix¶
Compute MI matrices¶
normalized_MI = compute_mutual_information_matrix(seq_kept)
MI = compute_mutual_information_matrix(seq_kept, normalize=False)
Compare MI versus SCA¶
The heatmap scales are adjusted to make the information visible¶
plt.figure(figsize=(12, 5))
for ih, (heatmap, title, vmm) in \
enumerate(zip([SCA_matrix, MI, normalized_MI],
['SCA', 'Mutual Information',
'Normalized Mutual Information'],
[[0, 1], [0, 0.8], [0, 0.2]])):
plt.subplot(1, 3, ih+1)
plt.imshow(heatmap, vmin=vmm[0], vmax=vmm[1], cmap='inferno')
plt.xlabel('residues', fontsize=10)
plt.ylabel(None)
plt.title('%s' % title)
plt.colorbar(shrink=0.4)
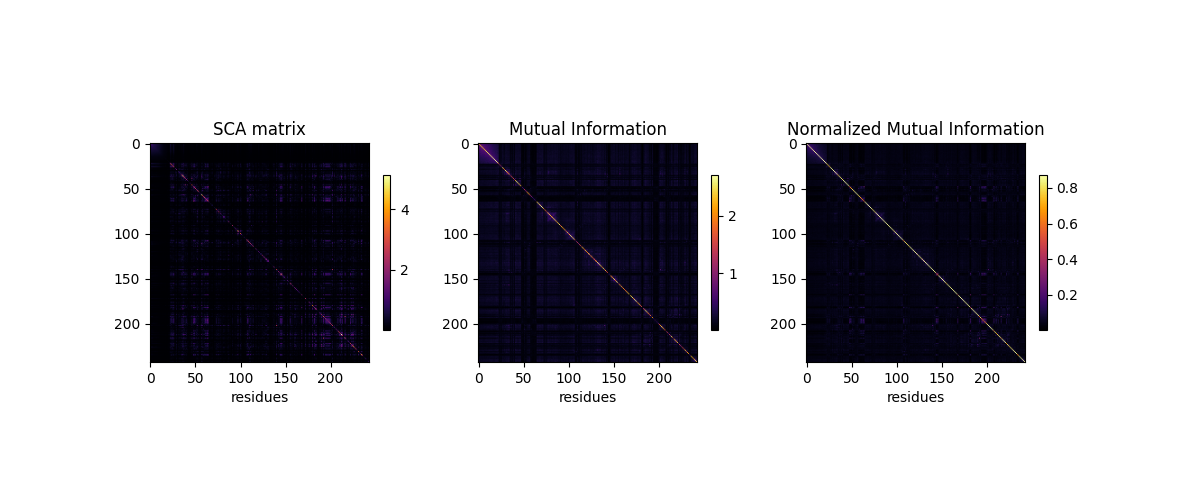
Relationships between SCA and MI’s¶
plt.figure(figsize=(12, 3))
plt.subplot(1, 3, 1)
plt.plot(np.triu(SCA_matrix, 1).flatten(),
np.triu(normalized_MI, 1).flatten(), 'o')
plt.xlabel('SCA')
plt.ylabel('Normalized Mutual Information')
plt.subplot(1, 3, 2)
plt.plot(np.triu(MI, 1).flatten(), np.triu(normalized_MI, 1).flatten(), 'o')
plt.xlabel('Mutual Information')
plt.ylabel('Normalized Mutual Information')
plt.subplot(1, 3, 3)
plt.plot(np.triu(SCA_matrix, 1).flatten(), np.triu(MI, 1).flatten(), 'o')
plt.xlabel('SCA')
plt.ylabel('Mutual Information')
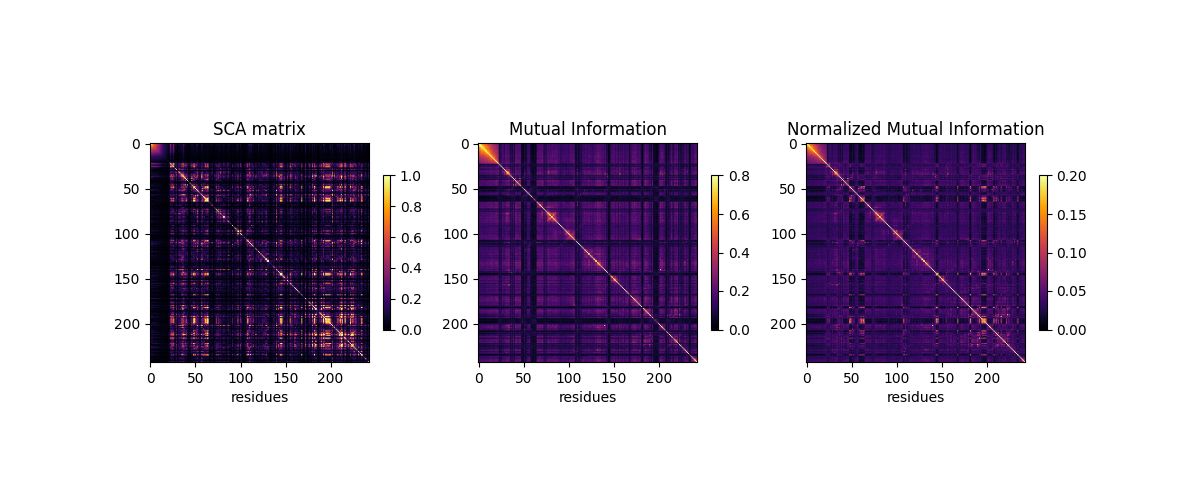
Text(769.1928104575164, 0.5, 'Mutual Information')
Relationship between coevolution metrics and conservation¶
plt.figure(figsize=(12, 3))
Di = compute_conservation(seq_kept)
for ih, (heatmap, title) in enumerate(zip([SCA_matrix, MI, normalized_MI],
['SCA', 'MI', 'normalized MI'])):
plt.subplot(1, 3, ih + 1)
cumul = np.sum(heatmap, axis=0) - np.diag(heatmap)
plt.plot(Di, cumul, 'o')
plt.xlabel('conservation')
if ih == 0:
plt.ylabel('cumulative score')
plt.title(title)
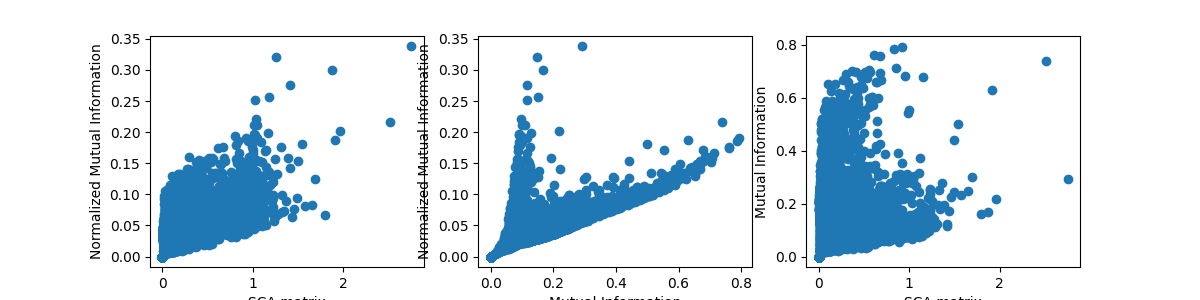
Total running time of the script: (0 minutes 6.593 seconds)